Research
Below are some of our present and past research projects.
Tool and Resource Development
We develop open source resources that can drive innovation and accelerate the discovery of novel therapeutic strategies for pediatric brain tumor research.
OpenPBTA: With Alex’s Lemonade Stand Foundation’s Childhood Cancer Data Lab, Dr. Rokita co-led the creation of the Open Pediatric Brain Tumor Atlas (OpenPBTA), an open-source reproducible analysis platform consisting of nearly 50 scalable modules to analyze pediatric brain tumor data.
OpenPedCan: The pan-cancer expansion counterpart of OpenPBTA is the Open Pediatric Cancer (OpenPedCan) Project. OpenPedCan became the foundational data layer of the NCI’s pediatric Molecular Targets Platform and is used to support clinical trial decision-making for the Pediatric Neuro-Oncology Consortium (PNOC), therapeutic development, and discovery and translational research.
AutoGVP: In collaboration with Dr. Sharon Diskin at CHOP, we created AutoGVP, a dockerized tool to assess germline variant pathogenicity based on current ACMG guidelines. We also created a pathogenicity preprocessing CAVATICA application to add ClinVar, InterVar, ANNOVAR, and AutoPVS1 annotations to the germline VCFs prior to running AutoGVP.
annoFuse: We created an R package, annoFuse and companion RShiny application, ShinyFuse to prioritize putative oncogenic fusions and visually explore fusion calls and breakpoints across a cohort.
Integrative Multi-omics Approach
Our research harnesses the power of integrative multi-omics to unravel the complexities of pediatric brain tumors. By combining data from genomics, transcriptomics, and proteomics, we uncover critical insights into the biology of these aggressive cancers.
RNA splicing mechanisms underlying oncogenesis: We investigate the role of RNA splicing in driving tumor formation and progression in pediatric brain tumors.
Identification of novel targets: We have developed a workflow to identify and prioritize splice-derived peptides as new candidate tumor-specific vulnerabilities, paving the way for personalized immunotherapeutic treatment strategies for pediatric brain tumor patients.
Germline risk variants: In collaboration with Dr. Sharon Diskin, our team characterizes patient and parental germlines to pinpoint pathogenic variants that predispose patients to brain tumor development, offering new insights into how these variants influence tumor biology.
Health Disparities
We are deeply passionate about exploring the impact of social determinants of health, race, ethnicity, and genetic-based ancestry on pediatric brain tumors. In collaboration with Dr. Cassie Kline, we recently identified significant associations between genetic ancestry superpopulations, prevalence of CNS tumor histologies and molecular subtypes, patient overall and event-free survival, and upfront treatment approaches.
Featured
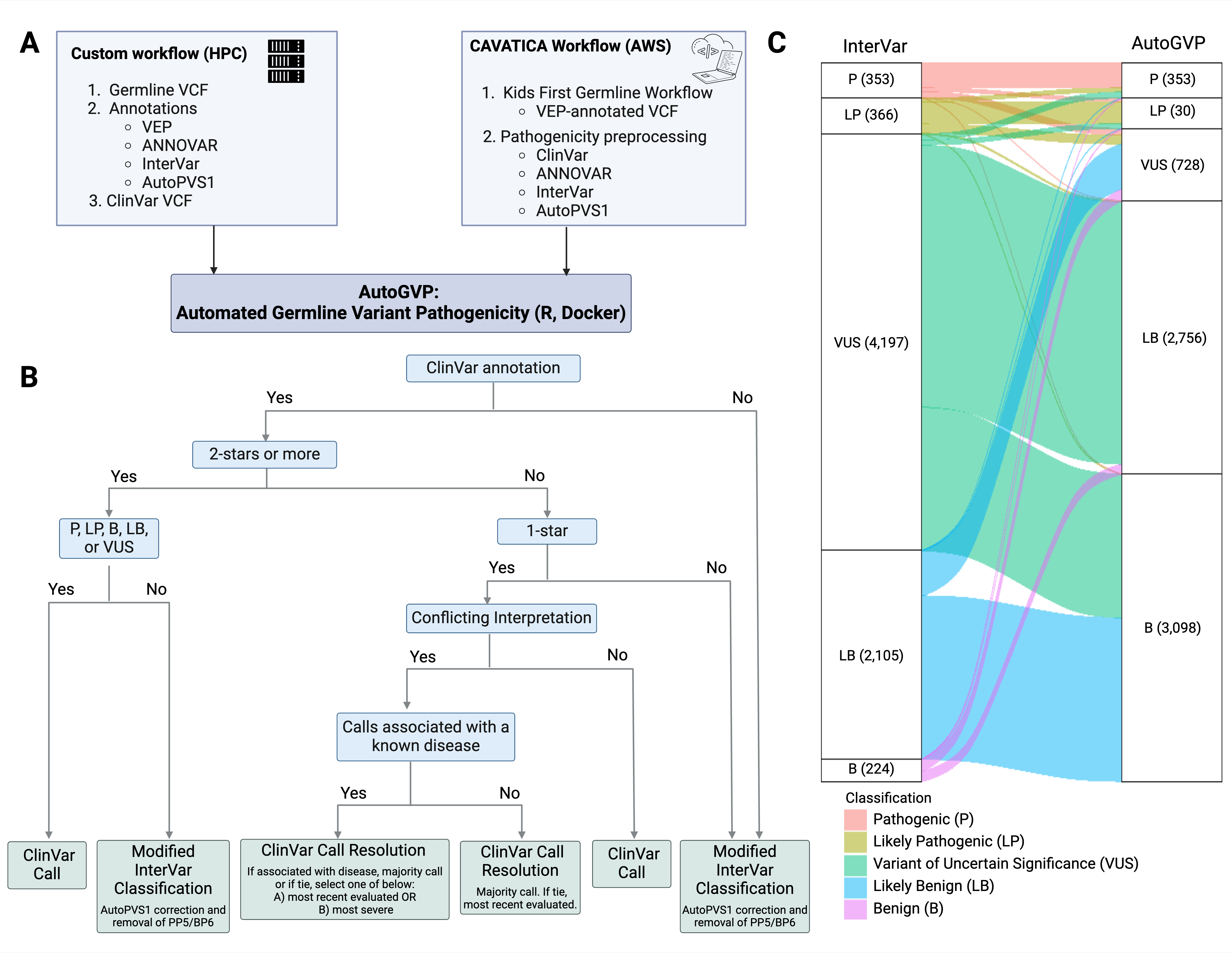
A dockerized workflow integrating ClinVar and InterVar germline sequence variant classification
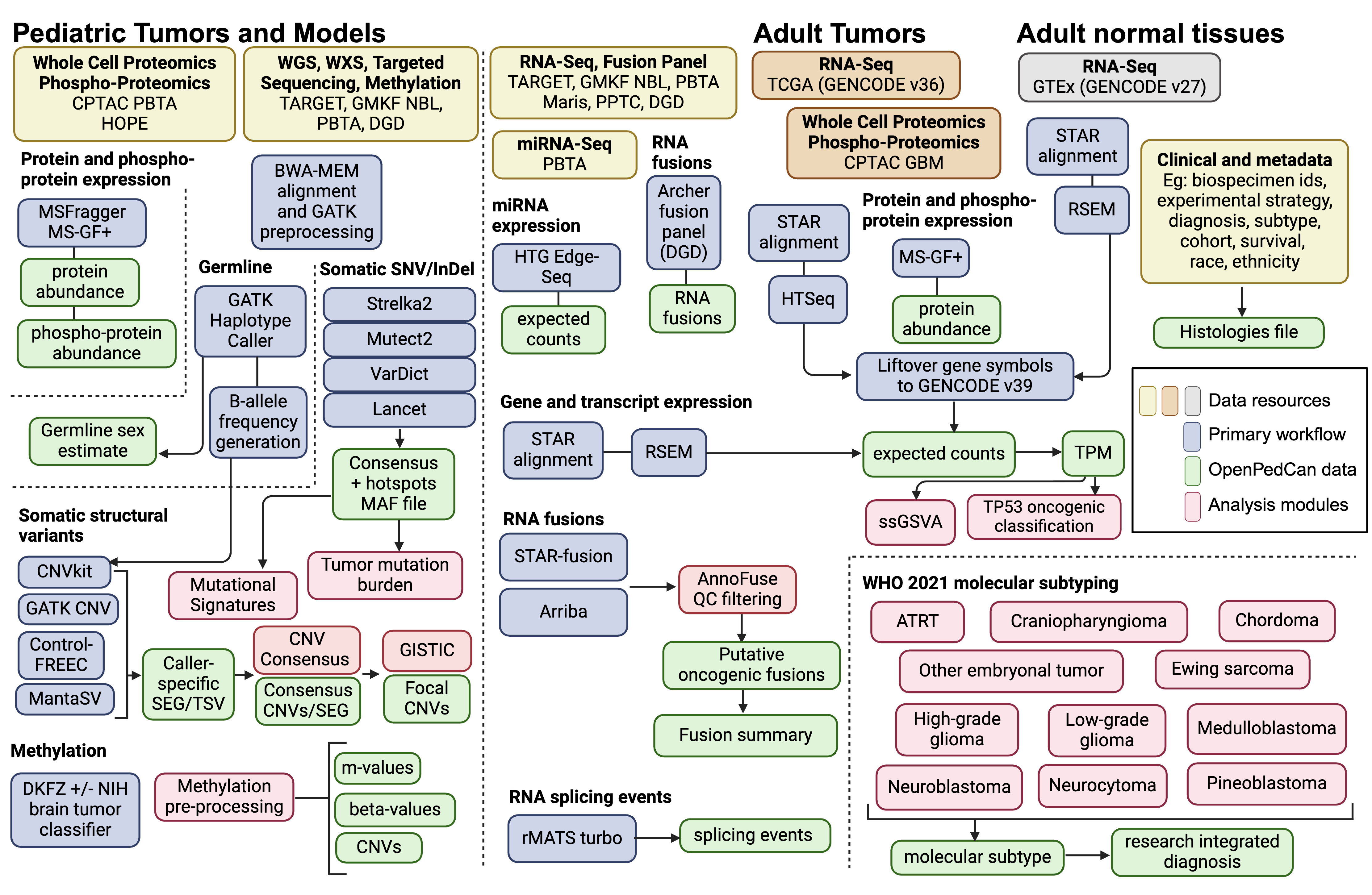
A harmonized open-source multi-omic dataset from > 6,000 pediatric cancer patients with > 7,000 tumor events across more than 100 histologies.
More
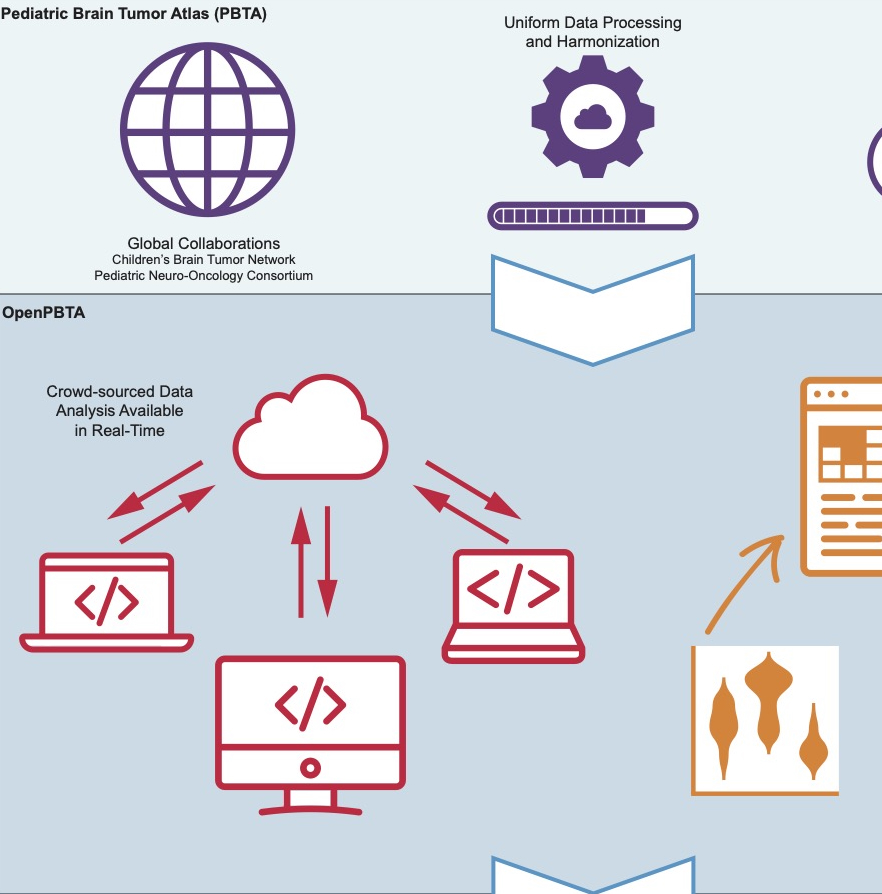
A global open science initiative characterizing over 1,000 pediatric brain tumors.
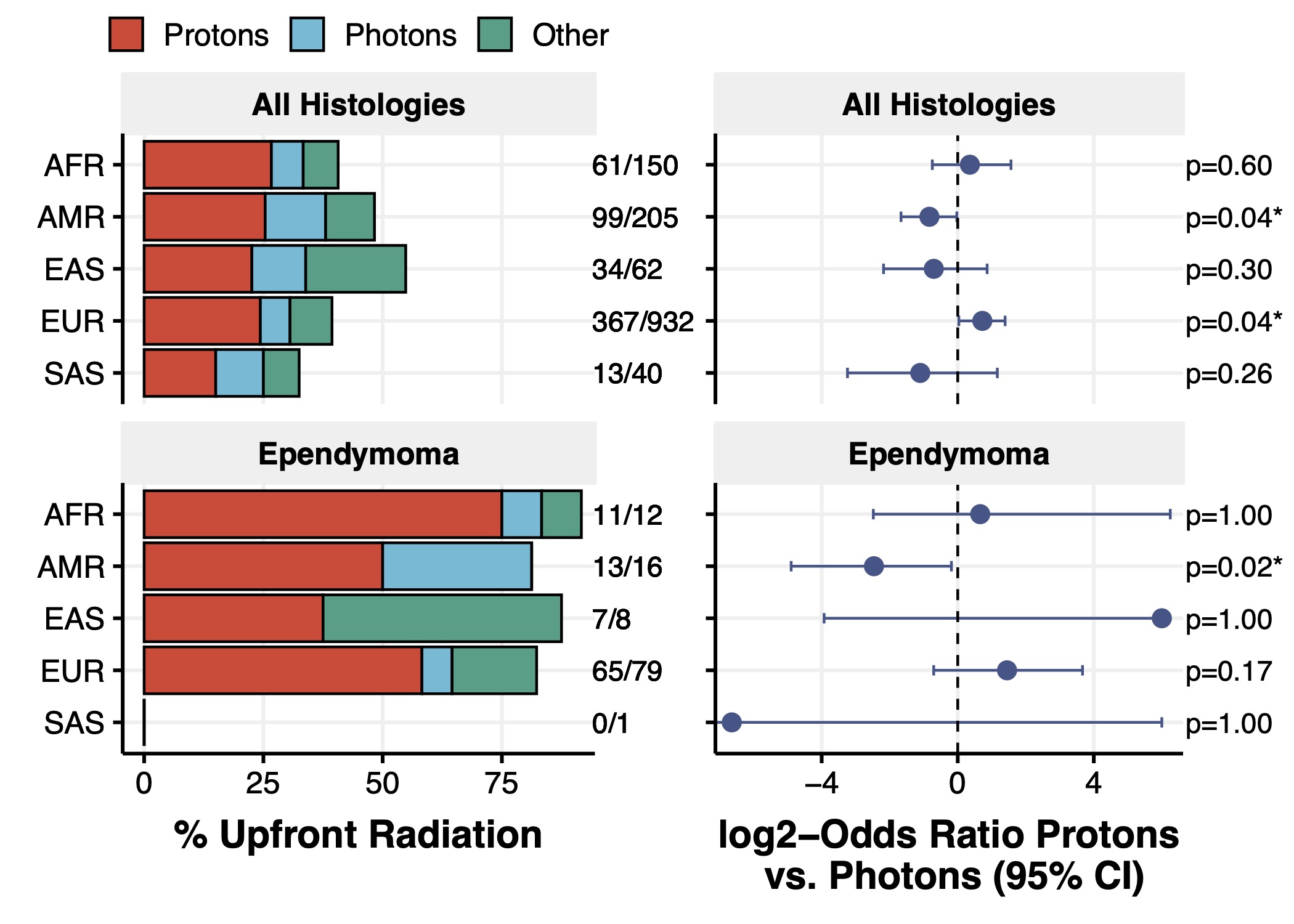
Distinct associations of genetic ancestry-based superpopulations exist within pediatric CNS tumor histologic and molecular subtypes and correlate with survival outcomes and treatment.
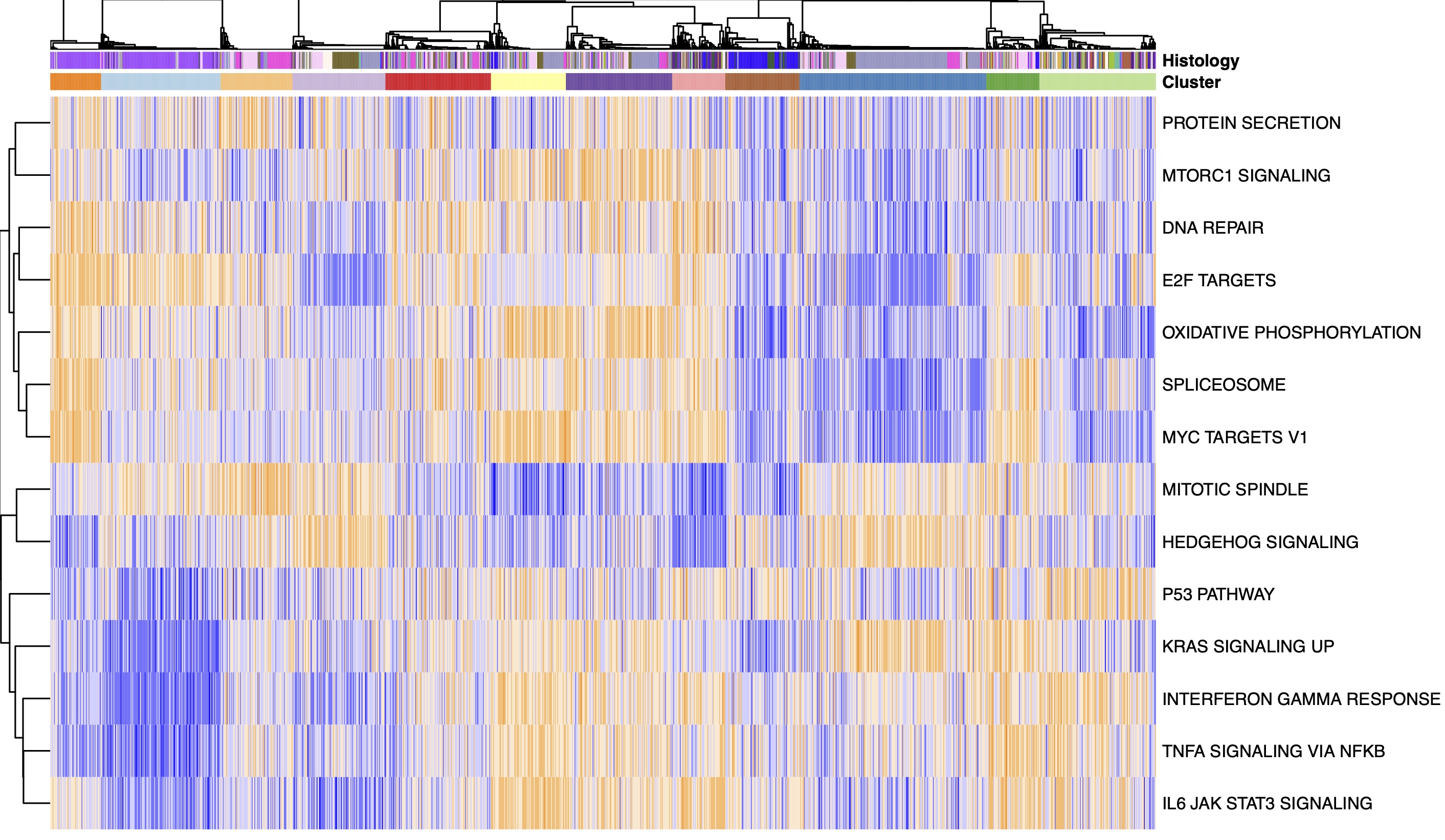
Characterization of aberrant splicing in pediatric central nervous system tumors reveals CLK1 as a candidate oncogenic dependency